VexGen DX is a Molecular Laboratory that utilizes Next-Gen DNA Sequencing to accurately identify all microbes within each sample. The extracted DNA from each sample is matched to the DNA sequence codes of the National data base of over 25,000+ known microbes of bacterial and fungal species with 99.9% accuracy. Conversely, we now know that culture methods can only grow 1% of all known microbes and have an alarming rate of negative or no growth results leaving veterinarians guessing as to what microbes are causing an infection and resisting treatment decisions.*
*Bjarnsholt T., “The role of bacterial biofilms in chronic infections.” US National Library of Medicine, APMIS Suppl. 2013 May;(136):1-51. doi: 10.1111/apm.12099., PubMed.gov, web.
Clinical Relevance of Next-Gen Sequencing (NGS)
What is Next-Generation Sequencing?
Chronic Infections, Biofilms & Next-Gen Sequencing
All planktonic or single cell microorganisms will eventually move to a biofilm state, which makes diagnosing the causative microbes by way of culture methods nearly impossible and leads to chronic and ongoing infections.
- NIH research shows that 80% of infections are caused by biofilms. Cultures fail to identify the microorganisms in a biofilm phenotype.”*
*Orthopaedic biofilm infections – Paul Stoodley,a Garth D. Ehrlich,b Parish P. Sedghizadeh,c Luanne Hall-Stoodley,d Mark E. Baratz,e Daniel T. Altman,e Nicholas G. Sotereanos,e John William Costerton,e and Patrick DeMeo.
Biofilms
To understand WHY cultures fail to correctly identify the causitive microbes we need to understand how biofilms form and work together to survive. Microbes have switched from an acute frontal attack by planktonic cells, to a strategy of biofilm growth and chronic attack on infected tissues, and the most serious long-term effect of this tactical change is that they evade detection by the classic methods of Medical Microbiology (culture method).
Patients will be treated more effectively when the identity of all bacteria in their tissues can be accurately determined.
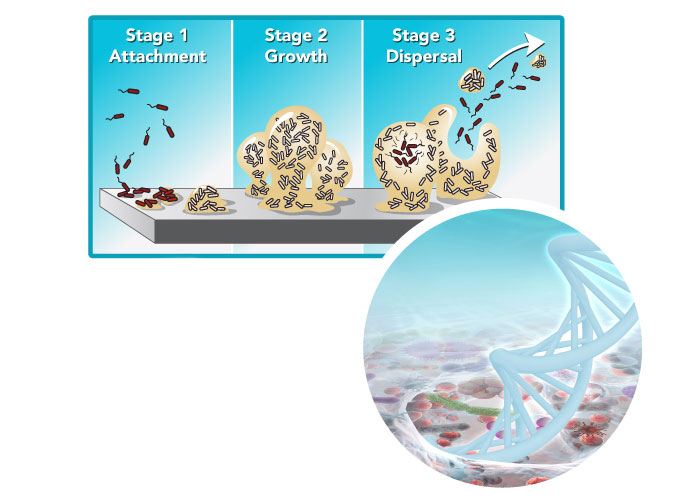
“To understand why cultures fail in correctly identifying the causative microbes we need to understand how biofilms form and work together to survive.”
Next-Gen Sequencing (NGS)
We live in the era of precise DNA-based forensics, and of whole genome sequencing, and patients will be best served when we mobilize these resources for the prompt and accurate diagnoses of bacterial disease. ONLY Next-Gen Sequencing (NGS) can detect all of the microbes that are living at a site of the infection.
Molecular Diagnostics Versus Any Microlab
Why a Microlab will not find or grow microbes that are detected with DNA Sequencing

How Could Contaminants Appear On My Lab Report?
Contamination could happen during the sampling process. For example, if proper skin prep is not done correctly, normal flora on the skin may attach itself to the needle while penetrating the skin for an aspiration sample a knee or shoulder. This should be considered if Staph Epi or P Acne are listed on the lab results. The area of the sample collection needs to be considered when reviewing your report to determine which microbes are to be treated.
Q: Could any of the microbes detected be contaminants?
A: Yes, there may be contaminants listed on your report and it is critical to follow good clinical practice when gathering samples to minimize the risk.
Healthy Microbiome Sites - Why They Matter
Sites That Have A Normal Microbiome?
Sinus Cavities, for example, have a microbiome of bacteria that work in harmony to keep the host healthy. When taking a sample it is crucial that the sample is taken from the site of the infection which means the sample should be collected from an area of inflammation.
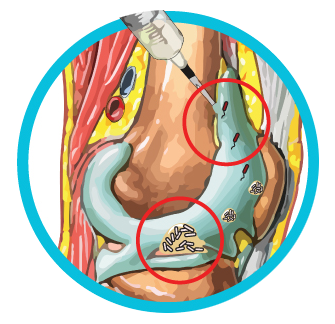
Sites That Do Not Have Normal Microbiomes:
Host areas that do not have a natural microbiome include those such as joints, wounds or lungs where bacteria and fungi should not be colonizing. In these cases, the NGS report is straightforward in detecting the bacteria and fungus that have been found at the site of collection. If the animal is not showing any other markers of infection then itis dealing with the species that have found their way to that part of the body. If there are any other laboratory tests, WBC, CRP, LE and/or clinical signs of inflammation and infection, then the NGS report provides the make-up of all microorganisms found at the site and should be considered as part of the overall picture in developing a treatment algorithm.
What To Do With The Lab Report Info?

Level I - PCR Test Result
qPCR Rapid Screening will detect 8-16+ microbes traditionally found or determined to cause a certain type of infection. Within 24hrs qPCR will identify the “usual suspects” at the scene of the infection but cannot identify any other species that may be there.
The test also reports on antibiotic resistance genes found in the sample in order to identify antibiotics that may not work for treatment of detected microbes.
The value of qPCR is limited to those 8-16+ identified microbes. However, all chronic infections contain many more species of microbes than those found by qPCR testing. VexGen DX has identified over 6,500 species of microorganisms in human samples to date! This is why NGS is so crucial for providing a comprehensive analysis of exactly which microbial species are at the site of an infection.
Level II – Next Gen Sequencing
To date, VexGen DX has over 6,500 microbial species in human samples. This level of detail is crucial in helping veterinarians treat the actual cause of an infection. There are often chronic infections that veterinarians stop treating due to “zero growth culture reports” and/or “no signs of improvement”. The NGS test provides veterinarians and animal owners with answers they could not get before. Everyday real-world human outcomes have changed lives by making the difference in clearing a 20-year sinus infection, in stopping years of UTI pain and suffering and in putting a stop to unnecessary amputations. It is not magic or a miracle, but rather the most comprehensive diagnostic tool a veterinarian will ever need to develop accurate and effective treatment regimens based on the accurate microbial data identified which is actually causing an infection. By relying only on a culture or qPCR report, there is a great possibility the veterinarian may be only aware of a fraction of the whole microbial picture. Next-Gen Sequencing provides the entire picture in a single affordable test.
Treatment Decisions Based on NGS
One School Of Thought
One school of thought to treat infection is to target the dominant species detected. By destroying the microbes present in the highest quantity, the biofilm “house” or community can collapse and the rest will be taken out under host pressure. Microlabs using traditional culture are also looking for a “dominant species” based on what will grow. NGS is identifying DNA evidence of the true dominant species at the host site, not just based on what grows best in agar.
Another School Of Thought
Another school of thought is to target all the species detected, not just the dominant species. This strategy has to take into consideration the use of multiple antimicrobials and the impact to the host. We know that multiple systemic antibiotics have consequences to the microbiome of the gut. We also know that these agents have side effects to take into consideration. For certain infections (sinus & wound, UTI ) a topical application of antimicrobials can provide a route of administration that provides the benefit of avoiding the impact to the host microbiome in the gut. Topical application of antimicrobials can avoid the toxicity and side effects of systemic delivery while delivering 1K to 5K times the concentration of antimicrobials directly on top of the site of infection. This high concentration would never have been achieved with oral or IV delivery and would provide greater penetration of biofilm EPS matrix.
“Topical application of antimicrobials can avoid the toxicity and side effects of systemic delivery while delivering 1K to 5K times the antimicrobial directly on top of the site of infection.”

Do I Have To Follow The Recommendations In The Report?
Cultures were designed to grow single cell / planktonic bacteria otherwise known as “professional pathogens”. Molecular diagnostics with NGS give you the complete picture. You make the decision as to what you will treat. The recommended antibiotics and antifungals are only offered as suggestions based on a research database where antimicrobials were shown to be effective against the species detected. It is up to the veterinarian to make use of the information in the report, along with other information gathered in the treatment of the animal, to decide on the best course of action. But the most significant factor NGS provides is accurate data of what is living inside the site of infection so you can be more confident in your treatment decisions and in their recovery and overall health.